Microbes that live in rice paddies, northern peat bogs and other previously unexpected environments are among the bacteria that can generate highly toxic methylmercury, researchers at Oak Ridge National Laboratory and the Smithsonian Environmental Research Center have learned.
Background:
Mercury is a global pollutant, released to the atmosphere through coal
burning and other industrial uses, and through natural processes. Methylmercury—the most dangerous form of mercury—damages the brain and
immune system and is especially harmful to developing embryos. Most
of the harm comes from methylmercury bioaccumulation, which is the
buildup of the element in tissue that occurs when moving up the food
chain. While it was known that certain special bacteria are able to transform inorganic mercury into methylmercury, the lack of a genetic marker for such biomethylation has prevented a clear understanding of mercury-methylating organism distribution in nature.
The new study:
The results of a new study, published in Environmental Science and Technology, explains why deadly methylated mercury is produced in areas where the neurotoxin's presence has puzzled researchers for decades.
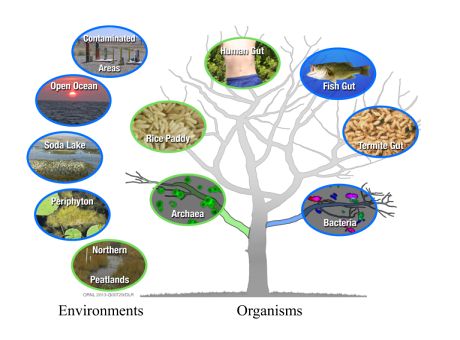
Newly discovered archaeal (green) and bacterial (blue) organisms that are free-living or within another organism on the tree of life, and their larger environments.
|
Elias and colleagues tested the hypothesis that a a specific gene cluster (hgcAB) is a marker for the capability of an organism to perform Hg methylation. Using this hypothesis they were able to show that organisms able to methylate mercury are much more phylogenetically and environmentally diverse than previously thought.
“We showed for the first time that many different types of bacteria are able to produce this potent neurotoxin,” Elias said. “The newly identified microbes include methane-producing organisms that live in rice paddies, anaerobic wastewater treatment plants, northern peat lands and possibly within our bodies.”
Elias and colleagues are testing a bacterium from the human intestine that they predict will also methylate mercury. Other bacteria able to transform inorganic into methylmercury include those used in biological dechlorination and metal treatment systems. All of these organisms are anaerobic, which means they grow in habitats without oxygen, including aquatic sediments and wetland soils.
By identifying these organisms, the researchers may have explained why methylmercury is accumulating in unexpected places. The discovery may also help clarify how methylmercury is produced in the open ocean, according to co-author Cindy Gilmour of the Smithsonian Environmental Research Center. Gilmour noted that the newly identified fermentative bacteria may be common in the low oxygen zones of the ocean where methylmercury production occurs. Ocean fish are the primary source of methylmercury in human diets worldwide.
Gilmour and Elias believe this work will have far-reaching implications for understanding the global mercury cycle. Scientists can now use the mercury-methylating organisms and their genes as global biomarkers and develop detection techniques to assess and eventually predict the extent of methylmercury production in a given environment.
“Based on the substantially expanded diversity of organisms and environments for methylation, a number of important habitats deserve further attention,” Gilmour said. “We now have a way to identify and track the abundance of mercury-methylating organisms within these environments, and we hope that the information can be used to devise strategies to minimize methylmercury production.”
Source: adapted from
ORNL The cited study:
The cited study:
Cynthia C. Gilmour, Mircea Podar, Allyson L Bullock, Andrew Mitchell Graham, Steven Brown, Anil C Somenahally, Alexander Johs, Richard Hurt, Kathryn L Bailey, Dwayne Elias,
Mercury methylation by novel microorganisms from new environments, Environ. Sci. Technol., Just Accepted Manuscript, 2013,
DOI: 10.1021/es403075t Related studies (newest first):
Related studies (newest first):
R.-Q. Yu, J.R. Reinfelder, M.E. Hines, T. Barkay,
Mercury methylation by the methanogen Methanospirillum hungatei, Appl. Environ. Microbiol. 2013, Published ahead of print August 9, 2013,
doi: 10.1128/AEM.01556-13
Haiyan Hu, Hui Lin, Wang Zheng, Stephen Tomanicek, Alexander Johs, Dwayne Elias, Liyuan Liang and Baohua Gu,
Oxidation and methylation of dissolved elemental mercury by anaerobic bacteria, Nature Geoscience, (2013)
doi: 10.1038/ngeo1894
J.M. Parks, A. Johs, M. Podar, R. Bridou, R.A. Hurt, S.D. Smith, S.J. Tomanicek, Y. Qian, S.D. Brown, C.C. Brandt, A.V. Palumbo, J.C. Smith, J.D. Wall, D.A. Elias, L. Liang,
The genetic basis for bacterial mercury methylation, Science, 339 (2013) 1332-1335.
DOI: 10.1126/science.1230667
H. Hsu-Kim, K.H. Kucharzyk, T. Zhang, M.A. Deshusses,
Mechanisms regulating mercury bioavailability for methylating microorganisms in the aquatic environment: A critical review, Environ. Sci. Technol., 47/6 (2013) 2441-2456.
DOI: 10.1021/es304370g
T. Zhang, B. Kim, C. Leyard, B.C. Reinsch, G.V. Lowry, M.A. Deshusses, H. Hsu-Kim,
Methylation of mercury by bacteria exposed to dissolved, nanoparticulate, and microparticulate mercuric sulfides, Environ. Sci. Technol., 46/13 (2012) 6950-6958.
DOI: 10.1021/es203181m
Sarah E. Rothenberg, Xinbin Feng,
Mercury cycling in a flooded rice paddy, J. Geophys. Res., 117 (2012) G03003.
doi:10.1029/2011JG001800
I. Tjerngren, T. Karlsson,
E. Björn, U. Skyllberg,
Potential Hg methylation and MeHg demethylation rates related to the nutrient status of different boreal wetlands, Biogeochem., 108/1-3 (2012) 335-350.
DOI: 10.1007/s10533-011-9603-1
A.M. Graham, G.R. Aiken, C.C. Gilmour,
Dissolved organic matter enhances
microbial mercury methylation under sulfidic conditions, Environ. Sci. Technol., 46/5 (2012) 2715-2723.
DOI: 10.1021/es203658f
R. Bridou,
M. Monperrus, P.R. Gonzalez, R. Guyoneaud,
D. Amouroux,
Simultaneous determination of mercury methylation and demethylation capacities of various sulfate-reducing bacteria using species-specific isotopic tracers, Environ. Toxicol. Chem., 30/2 (2011) 337-344.
DOI: 10.1002/etc.395
S. Hamelin, M. Amyot, T. Barkay, Y.P. Wang, D. Planas,
Methanogens: Principal methylators of mercury in lake periphyton, Environ. Sci. Technol., 45/18 (2011) 7693-7700.
DOI: 10.1021/es2010072
S.S. Gbondo-Tugbawa, J.A. McAlear, C.T. Driscoll, C.W. Sharpe,
Total and methyl mercury transformations and mass loadings within a wastewater treatment plant and the impact of the effluent discharge to an alkaline hypereutrophic lake, Water Res., 44/9 (2010) 2863-2875.
DOI: 10.1016/j.watres.2010.01.028
E.G. Malcolm, J.K. Schaefer, E.B. Ekstrom, C.B. Tuit, A. Jayakumar, H. Park, B.B. Ward, F.M.M. Morel,
Mercury methylation in oxygen deficient zones of the oceans: No evidence for the predominance of anaerobes, Mar. Chem., 122/1-4 (2010) 11-19.
DOI: 10.1016/j.marchem.2010.08.004
M. Ranchou-Peyruse,
M. Monperrus, R. Bridou, R. Duran,
D. Amouroux, J.C. Salvado, R. Guyoneaud,
Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: Implications for environmental studies, Geomicrobiol. J., 26/1 (2009) 1-8.
DOI: 10.1080/01490450802599227
E.M. Sunderland, D.P. Krabbenhoft, J.W. Moreau, S.A. Strode, W.M. Landing,
Mercury sources, distribution, and bioavailability in the North Pacific Ocean: Insights from data and models, Global Biogeochem. Cycles, 23 (2009) GB2010.
DOI: 10.1029/2008GB003425
U. Skyllberg,
Competition among thiols and inorganic sulfides and polysulfides for Hg and MeHg in wetland soils and sediments under suboxic conditions: Illumination of controversies and implications for MeHg net production, J. Geophys. Res. Biogeosci., 113 (2008) G00C03.
DOI: 10.1029/2008JG000745
C.P.J. Mitchell, C.C. Gilmour,
Methylmercury production in a Chesapeake Bay salt marsh, J. Geophys. Res. -Biogeosci., 113 (2008) G00C04.
DOI: 10.1029/2008JG000765

C.P.J. Mitchell, B.A. Branfireun, R.K. Kolka,
Assessing sulfate and carbon controls on net methylmercury production in peatlands: An in situ mesocosm approach, Appl. Geochem., 23/3 (2008) 503-518.
DOI: 10.1016/j.apgeochem.2007.12.020
J.L. Kirk, V.L.S. Louis,
H. Hintelmann, I. Lehnherr, B. Else, L. Poissant,
Methylated mercury species in marine waters of the Canadian High and Sub Arctic, Environ. Sci. Technol., 42/22 (2008) 8367-8373.
DOI: 10.1021/es801635m
E.J. Fleming, E.E. Mack, P.G. Green, D.C. Nelson,
Mercury methylation from unexpected sources: Molybdate-inhibited freshwater sediments and an iron-reducing bacterium, Appl. Environ. Microbiol., 72/1 (2006) 457-464.
DOI: 10.1128/AEM.72.1.457-464.2006 
E.J. Kerin, C.C. Gilmour, E. Roden, M.T. Suzuki, J.D. Coates, R.P. Mason,
Mercury methylation by dissimilatory iron-reducing bacteria, Appl. Environ. Microbiol., 72/12 (2006) 7919-7921.
DOI: 10.1128/AEM.01602-06
M.S. Gustin, P.V. Chavan, K.E. Dennett, S. Donaldson, E. Marchand, G. Fernanadez,
Use of constructed wetlands with four different experimental designs to assess the potential for methyl and total Hg outputs, Appl. Geochem., 21/11 (2006) 2023-2035.
DOI: 10.1016/j.apgeochem.2006.08.012
E.B. Ekstrom, F.M.M. Morel, J.M. Benoit,
Mercury methylation independent of the acetyl-coenzyme A pathway in sulfate-reducing bacteria, Appl. Environ. Microbiol., 69/9 (2003) 5414-5422.
DOI: 10.1128/AEM.69.9.5414-5422.2003 
J.M. Benoit, C.C. Gilmour, R.P. Mason,
Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3), Appl. Environ. Microbiol., 67/1 (2001) 51-58.
DOI: 10.1128/AEM.67.1.51-58.2001
C.R. Hammerschmidt, W.F. Fitzgerald,
Formation of artifact methylmercury during extraction from a sediment reference material, Anal. Chem., 2001, 73/24 (2001) 5930-5936.
DOI: 10.1021/ac010721w
J.K. King, J.E. Kostka, M.E. Frischer, F.M. Saunders,
Sulfate-reducing bacteria methylate mercury at variable rates in pure culture and in marine sediments, Appl. Environ. Microbiol., 66/6 (2000) 2430-2437.
DOI: 10.1128/AEM.66.6.2430-2437.2000
S.C. Choi, R. Bartha, R. C
obalamin-mediated mercury methylation by Desulfovibrio desulfuricans LS, Appl. Environ. Microbiol., 59/1 (1993) 290-295.
available at: http://aem.asm.org/content/59/1/290.long Related EVISA Resources
Related EVISA Resources Link database: Mercury exposure through the diet
Link database: Mercury exposure through the diet Link database: Mercury and human health
Link database: Mercury and human health Link database: Environmental cycling of mercury
Link database: Environmental cycling of mercury Link database: Toxicity of Organo-mercury compounds
Link database: Toxicity of Organo-mercury compounds Link database: Research projects related to organo-mercury compounds
Link database: Research projects related to organo-mercury compounds Related EVISA News
Related EVISA News
 August 6, 2013: Bacterial methylation of mercury not only starting from oxidized mercury
August 6, 2013: Bacterial methylation of mercury not only starting from oxidized mercury
 February 8, 2013: ORNL scientists solve mystery about mercury methylation
February 8, 2013: ORNL scientists solve mystery about mercury methylation  June 17, 2012: Factors Affecting Methylmercury Accumulation in the Food Chain
June 17, 2012: Factors Affecting Methylmercury Accumulation in the Food Chain  October 9, 2006: Linking atmospheric mercury to methylmercury in fish
October 9, 2006: Linking atmospheric mercury to methylmercury in fish  February 17, 2006: Study shows link between clear lakes and methylmercury contamination in fish
February 17, 2006: Study shows link between clear lakes and methylmercury contamination in fish last time modified: September 15, 2013